
Background removal
Background subtraction is essential for effective quantification. For example, AzureSpot has five automatic background options and three manual options.

> If the background is uneven, use Rolling Ball. A disc of a specified radius "rolls" under the lane profile, averaging the signals to produce a smooth, small change. The larger the radius, the smoother the scrolling. 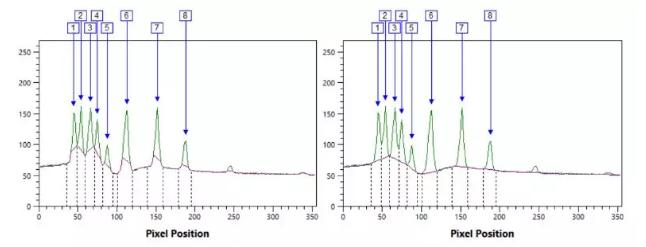
> If the end of the outline is not much different from the rest of the outline, use the Rubber Band. This method is like stretching a rubber band under the contour of a lane. Use this method with caution, find the lowest point at the edge of the lane, draw a slightly different lane, and the value will change. Usually, this calculation method is difficult to do repeated analysis.
> If you have performed strip detection, you can use Valley to Valley, which uses the edges of each strip to determine and subtract the background. It is important to enter the maximum slope to avoid identifying the edges between overlapping strips as the background.
> The Minimum Profile method is to set the minimum value to Background > LaneEdge Subtraction to use the lower of the two values ​​for each lane edge as the background.
Normalization AzureSpot can perform normalization in three ways. First, it sets the normalization of specific bands in the lanes in the standard gel. In multiple gels, it can be normalized with housekeeping proteins or total proteins.
Total protein normalization (TPN) is becoming more common as it has higher accuracy and adaptability to quantitative Western blotting. If you have verified that the expression of your housekeeping protein has not changed, then it may be appropriate. Otherwise, TPN is preferred.
The membrane is stained with a total protein stain (e.g., AzureRed fluorescent total protein stain) prior to post-transfer immunoassay. This method is not dependent on antibodies and takes into account changes in sample protein loading and transfer efficiency.
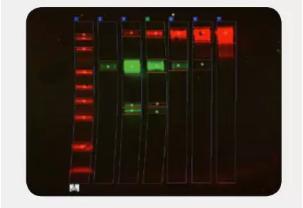
Don't forget to use the dilution series to confirm the linear detection range, which ensures that the signal strength is proportional to the sample size.
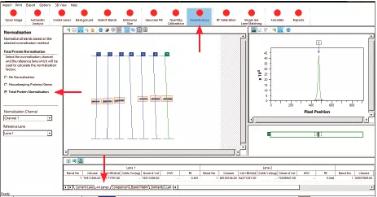
AzureSpot performs normalization. The purple line is always the background, and the green line above will be the signal. First select a lane as a reference lane (or standardized standard). For each lane, the normalization factor is calculated as the gray value of all the bands in the lane divided by the gray values ​​of all the bands in the reference lane. The normalized values ​​in all channels are then calculated, ie the original gray value of the strip divided by the normalization factor.
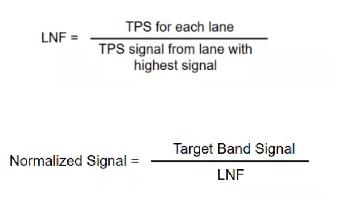
(LNF: normalized factor TPS: total protein staining)
The same amount of protein sample was added to the entire gel. Protein concentration determination can help determine if the concentration needs to be adjusted.
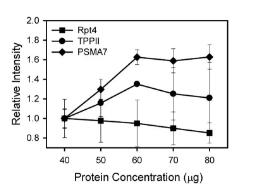
Examples of signal intensity for different proteins at different concentrations (Ghosh, et al. 2014
Make sure your sample load is in a linear dynamic range before normalization, which varies by concentration
With effective background subtraction and total protein normalization, you will be able to perform an effective quantitative analysis of phosphorylated proteins.
Pared Noodle,Wheat Ramen Noodles,Best Fresh Ramen Noodles,Instant Dry Noodles
Zhejiang Shanying Trading Co.,Ltd. , https://www.shanyingtrading.com