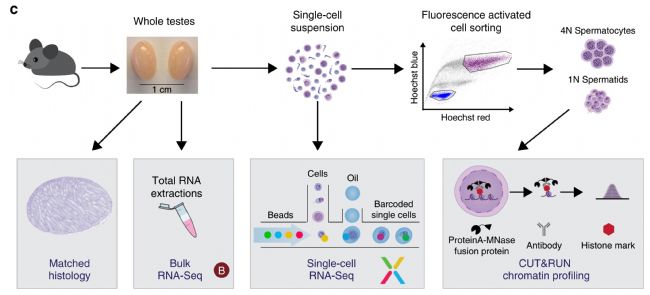
2. Single-cell sequencing for immune research, multi-point flowering (1) Memory CD4+ T cells
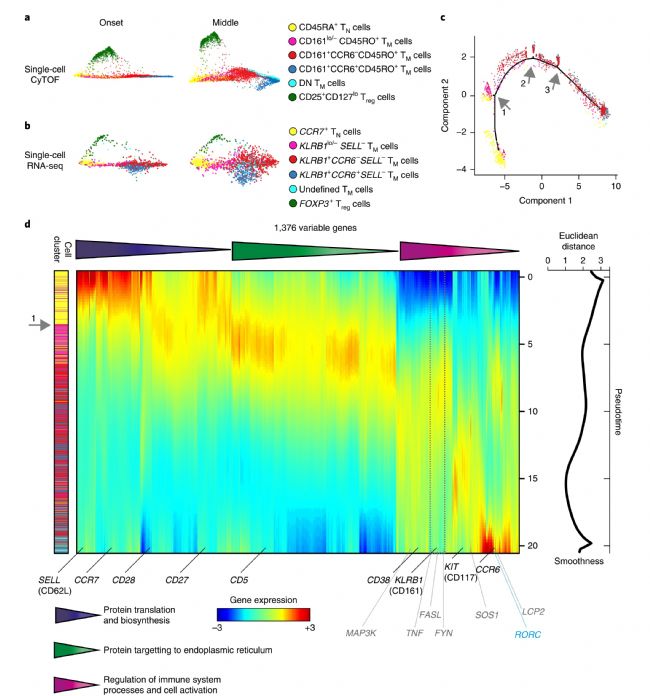
(3) Stem cell memory-like T cell production process tracking
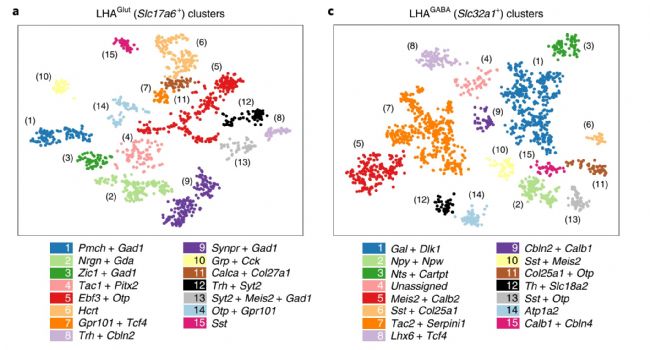
4. Single-cell transcriptome sequencing emerges in the field of plants
Compared to single-cell studies in animals, the single-cell study of plants is difficult due to the cell wall characteristics of plants, but following the single-cell transcriptomics study of Arabidopsis root tissue in February [8], this month in "Developmental A single-cell transcriptomics study based on Arabidopsis root tissue was published in Cell [9]. Researchers from the Center for Plant Molecular Biology at the University of Tübingen in Germany used 10X Genomics single-cell transcriptome sequencing to identify Arabidopsis root tissue cells. This map provides detailed temporal and spatial information, identifying all major cell types, including rare cells in the quiescent center (QC cells), revealing key developmental regulators and downstream pathways in the process of cell fate transformation into unique cell shapes and functions. gene. Through quasi-time series analysis, the study depicts the fine-grained trajectories from cells from niche to differentiation and the major regulatory transcription factors. At about the same time, researchers at the University of Washington's Genomics Science Center also used Arabidopsis root tissue single-cell sequencing studies to publish Arabidopsis root map results in the plant's top journal, Plant Cell [10]. In this study, in addition to the root cell classification similar to the above two articles, heat stress treatment was used to reveal the heterogeneity of response within the cell under abiotic stress. This study of surface single-cell transcriptomics studies has broad prospects in plant development and physiology.
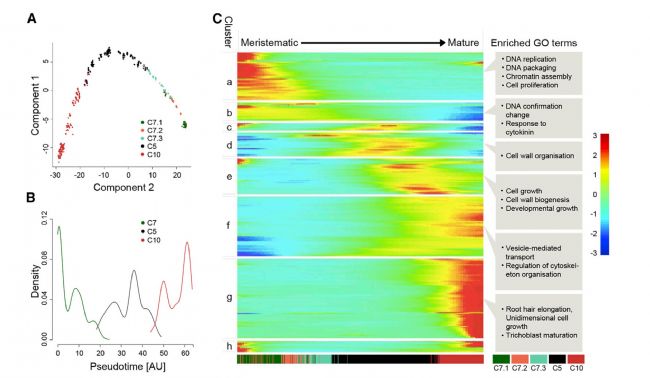
Figure 5 Gene change heat map from root cell to cell maturation process
5. Advances in single-cell research methods <br> In addition to the above research on single-cell transcriptomics, methodological studies based on single-cellomics analysis have also been published this month, including the Nature Method for cellular component analysis. CPM method [11], Nucleic Acids Research on cell-specific network construction based on single-cell transcriptome data [12], Genome Biology on low-transcription-level cell recognition based on droplet single-cell transcriptome data ( The development of the EmptyDrops software [13], the development of the Palantir algorithm for the identification of cell likelihoods in Nature Biotechnology [14] and the research on the optimization of tSNE algorithm in the Nature Method [15]. These studies have provided a powerful tool for the further development of future single-cellomics research.
In order to facilitate the single-cellomics research of domestic researchers, Shanghai Biochip Co., Ltd. (SBC) introduced the 10x Genomics single-cellomics detection platform in 2018, which provides an integrated solution from sample to analysis for researchers. We welcome all scientific research friends to carry out in-depth communication and extensive cooperation with us.
- Guo, J., et al., The adult human testis transcriptional cell atlas. Cell Res, 2018. 28 (12): p. 1141-1157.
- Sohni, A., et al., The Neonatal and Adult Human Testis Defined at the Single-Cell Level. Cell Rep, 2019. 26 (6): p. 1501-1517 e4.
- Ernst, C., et al., Staged developmental mapping and X chromosome transcriptional dynamics during mouse spermatogenesis. Nat Commun, 2019. 10 (1): p. 1251.
- Li, N., et al., Memory CD4(+) T cells are generated in the human fetal intestine. Nat Immunol, 2019. 20 (3): p. 301-312.
- Miller, BC, et al., Subsets of exhausted CD8(+) T cells differentially mediate tumor control and respond to checkpoint blockade. Nat Immunol, 2019. 20 (3): p. 326-336.
- Good, Z., et al., Proliferation tracing with single-cell mass cytometry optimizes generation of stem cell memory-like T cells. Nat Biotechnol, 2019. 37 (3): p. 259-266.
- Mickelsen, LE, et al., Single-cell transcriptomic analysis of the lateral hypothalamic area reveals molecularly distinct populations of inhibitory and excitatory neurons. Nat Neurosci, 2019. 22 (4): p. 642-656.
- Ryu, KH, et al., Single-cell RNA sequencing resolves molecular relationships among individual plant cells. Plant Physiol, 2019.
- Denyer, T., et al., Spatiotemporal Developmental Trajectories in the Arabidopsis Root Revealed Using High-Throughput Single-Cell RNA Sequencing. Dev Cell, 2019. 48 (6): p. 840-852 e5.
- Jean-Baptiste, K., et al., Dynamics of gene expression in single root cells of A. thaliana. Plant Cell, 2019.
- Frishberg, A., et al., Cell composition analysis of bulk genomics using single-cell data. Nat Methods, 2019.
- Dai, H., et al., Cell-specific network constructed by single-cell RNA sequencing data. Nucleic Acids Res, 2019.
- Lun, ATL, et al., EmptyDrops: distinguishing cells from empty droplets in droplet-based single-cell RNA sequencing data. Genome Biol, 2019. 20 (1): p.
- Setty, M., et al., Characterization of cell fate probabilities in single-cell data with Palantir. Nat Biotechnol, 2019.
- Linderman, GC, et al., Fast interpolation-based t-SNE for improved visualization of single-cell RNA-seq data. Nat Methods, 2019. 16 (3): p. 243-245.
Real-Time Fluorescence Quantitative PCR Systerm
This RT-PCR system is a necessary equipment for quantitative analysis of 2019-nCoV in big quantity during the COVID-19 pandemic, and this real-time PCR system can also be widely used in various fields such as scientific research, clinical detection and diagnosis, quality and safety testing, and forensic applications.
real time pcr equipment,real time pcr system,real time pcr device,real time pcr machine,rt pcr instrument
Shenzhen Uni-medica Technology Co.,Ltd , https://www.unimedicadevice.com